Enzyme is a plugin that performs automatic differentiation (AD) of statically analyzable LLVM.
Enzyme can be used by calling __enzyme_autodiff on a function to be differentiated as shown below.
Running the Enzyme transformation pass then replaces the call to __enzyme_autodiff with the gradient of its first argument.
double foo(double);
double grad_foo(double x) {
return __enzyme_autodiff(foo, x);
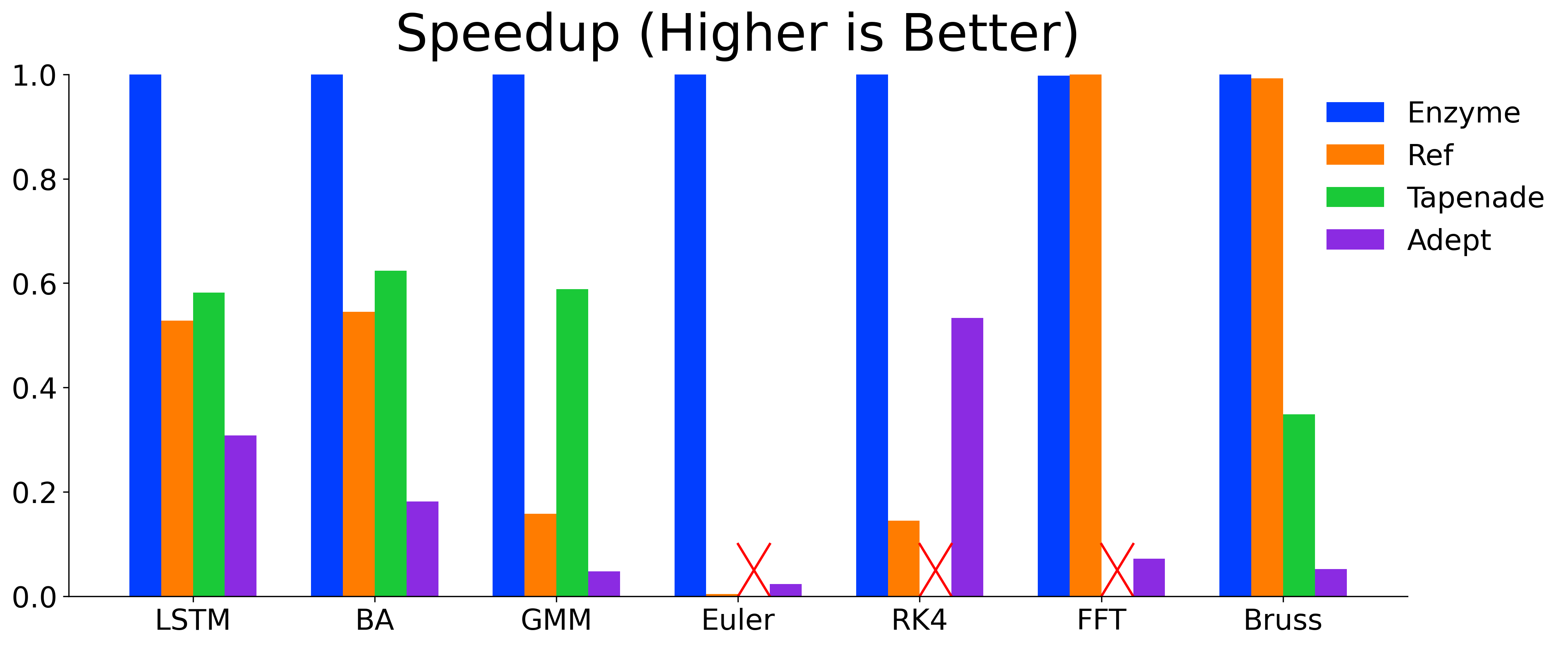
}Enzyme is highly-efficient and its ability to perform AD on optimized code allows Enzyme to meet or exceed the performance of state-of-the-art AD tools.
Detailed information on installing and using Enzyme can be found on our website: https://enzyme.mit.edu.
A short example of how to install Enzyme is below:
cd /path/to/Enzyme/enzyme
mkdir build && cd build
cmake -G Ninja .. -DLLVM_DIR=/path/to/llvm/lib/cmake/llvm -DLLVM_EXTERNAL_LIT=/path/to/lit/lit.py
ninja
To get involved or if you have questions, please join our mailing list.
If using this code in an academic setting, please cite the following:
@incollection{enzymeNeurips,
title = {Instead of Rewriting Foreign Code for Machine Learning, Automatically Synthesize Fast Gradients},
author = {Moses, William S. and Churavy, Valentin},
booktitle = {Advances in Neural Information Processing Systems 33},
year = {2020},
note = {To appear in},
}
Julia bindings for Enzyme are available here