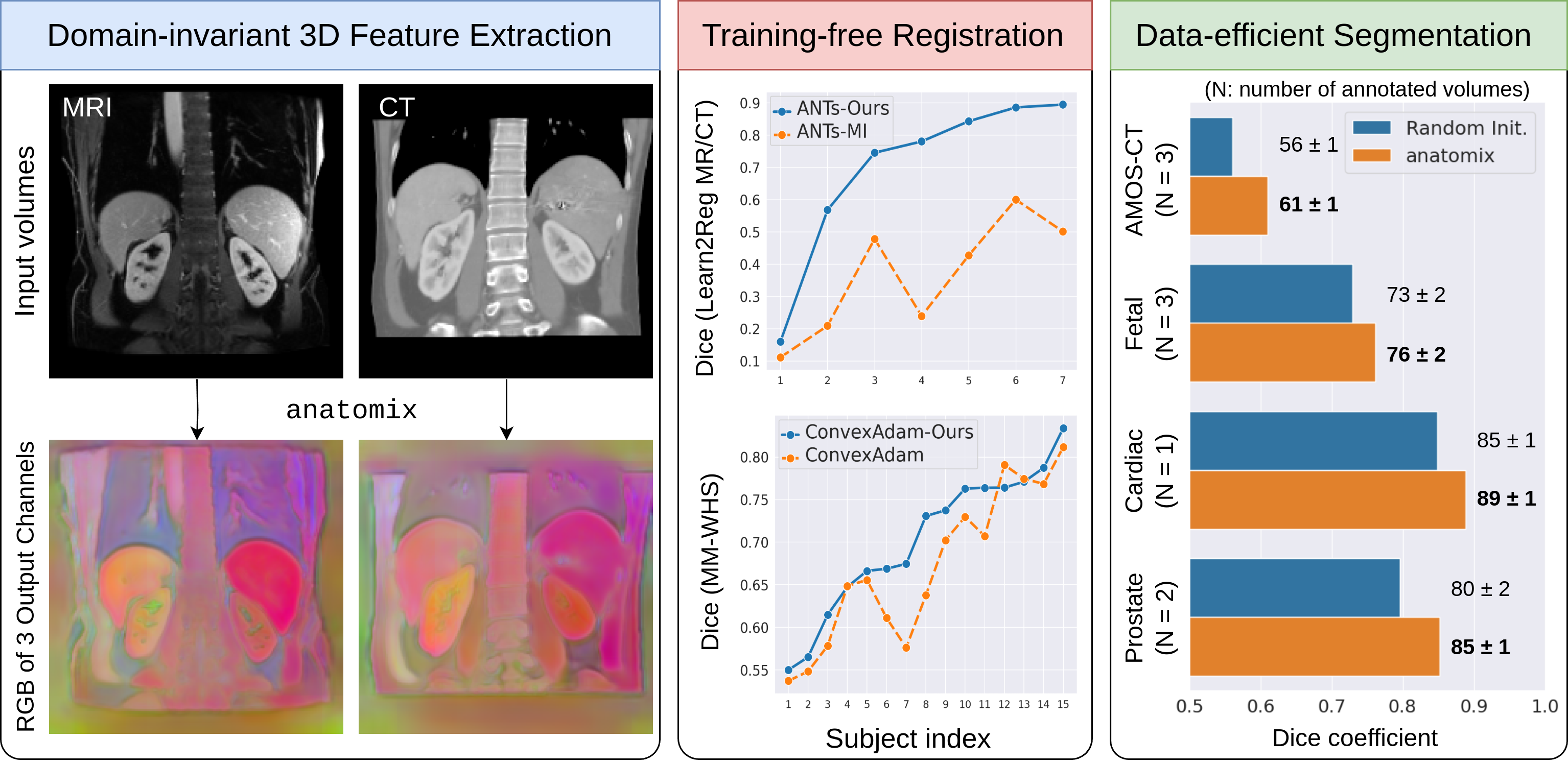
anatomix is a general-purpose feature extractor for 3D volumes. For any new biomedical dataset or task,
- Its out-of-the-box features are invariant to most forms of nuisance imaging variation.
- Its out-of-the-box weights are a good initialization for finetuning when given limited annotations.
Without any dataset or domain specific pretraining, this respectively leads to:
- SOTA 3D training-free multi-modality image registration
- SOTA 3D few-shot semantic segmentation
How? It's weights were contrastively pretrained on wildly variable synthetic volumes to learn approximate appearance invariance and pose equivariance for images with randomly sampled biomedical shape configurations with random intensities and artifacts.
Note: This repo was recently bumped up to PyTorch 2.8.0 and Python 3.11. If you find any bugs or changes in performance, please open an issue.
anatomix is just a pretrained UNet! Use it for whatever you like.
import torch
from anatomix.model.network import Unet
model = Unet(
dimension=3, # Only 3D supported for now
input_nc=1, # number of input channels
output_nc=16, # number of output channels
num_downs=4, # number of downsampling layers
ngf=16, # channel multiplier
)
model.load_state_dict(
torch.load("./model-weights/anatomix.pth"),
strict=True,
)See how to use it on real data for feature extraction or registration in this tutorial. Or if you want to finetune for your own task, check out this tutorial instead.
(If your task involves brains, you might benefit by using the anatomix+brains.pth weights instead, where we synthesize training volumes using both our synthetic label ensembles and real brain labels.)
All scripts will require the anatomix environment defined below to run.
conda create -n anatomix python=3.11
conda activate anatomix
git clone https://github.com/neel-dey/anatomix.git
cd anatomix
pip install -e .
Or install the dependencies manually:
conda create -n anatomix python=3.11
conda activate anatomix
pip install numpy nibabel scipy scikit-image nilearn h5py matplotlib torch==2.8.0 tensorboard tqdm monai torchio SimpleITK
This repository contains:
- Model weights for
anatomix(and its variants) - Scripts for 3D nonrigid registration using
anatomixfeatures - Scripts for finetuning
anatomixweights for semantic segmentation.
Each subfolder (described below) has its own README to detail its use.
root-directory/
│
├── model-weights/ # Pretrained model weights
│
├── pretraining/ # Pretraining code and scripts
│
├── synthetic-data-generation/ # Scripts to generate synthetic training data
│
├── anatomix/model/ # Model definition and architecture
│
├── anatomix/registration/ # Scripts for registration using the pretrained model
│
├── anatomix/segmentation/ # Scripts for fine-tuning the model for semantic segmentation
│
└── README.md # This fileThe current repository is just an initial push. It will be refactored and some quality-of-life and core aspects will be pushed as well in the coming weeks. These include:
- Contrastive pretraining code
- Colab 3D feature extraction tutorial
- Colab 3D multimodality registration tutorial
- Colab 3D few-shot finetuning tutorial
- General-purpose registration interface with FireANTs
- Dataset-specific modeling details for paper
If you find our work useful, please consider citing:
@misc{dey2024learninggeneralpurposebiomedicalvolume,
title={Learning General-Purpose Biomedical Volume Representations using Randomized Synthesis},
author={Neel Dey and Benjamin Billot and Hallee E. Wong and Clinton J. Wang and Mengwei Ren and P. Ellen Grant and Adrian V. Dalca and Polina Golland},
year={2024},
eprint={2411.02372},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2411.02372},
}
Portions of this repository have been taken from the Contrastive Unpaired Translation and ConvexAdam repositories and modified. Thanks!